Repeated Measures ANOVA (rma)
- equivalent of the one-way ANOVA, but for related, not independent groups, \item in that sense it is the extension of the dependent t-test: he same people are being measured more than once on the same dependent variable
- here we will look at the simplest design of rma
Statistical Model
- remember in basic one-way Anova
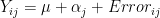
- error was assumed to have variance of
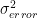
- measurements and therefore the errors were independed
Statistical Model
- in repeated measurement Anova
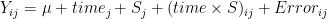
- assumes still that errors have the same variances and
- covariances are the same
Repeated Measures ANOVA (rma)
- main hypothesis of rma:
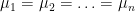
- here we will look at the simplest design of rma
- in this design the within group variance from above is splitted in variance caused by subject variability and the error variance
Repeated Measures ANOVA (rma)
So we have one more sum of sqares:
- SS_Time
- SS_within
- SS_Subject
- SS_error
Example
1 > xx
2 subject t1 t2 t3
3 1 1 45 50 55
4 2 2 42 42 45
5 3 3 36 41 43
6 4 4 39 35 40
7 5 5 51 55 59
8 6 6 44 49 56
9 > n <- 6
10 > k <- 3
11 > require(reshape2)
12 > xx <- melt(xx,id.vars = "subject")
13 > ## Sum of Squares time
14 > (tmp <- aggregate(value ~ variable,FUN = "mean",data=xx))
15 variable value
16 1 t1 42.83333
17 2 t2 45.33333
18 3 t3 49.66667
19 > (sstime <- sum(6*(tmp$value - mean(xx$value))**2))
20 [1] 143.4444
21 > ## sum of squares within
22 > (xx <- xx %>% group_by(variable) %>% mutate(gr.mean=mean(value)))
23 subject variable value gr.mean
24 1 1 t1 45 42.83333
25 2 2 t1 42 42.83333
26 3 3 t1 36 42.83333
27 4 4 t1 39 42.83333
28 5 5 t1 51 42.83333
29 6 6 t1 44 42.83333
30 7 1 t2 50 45.33333
31 8 2 t2 42 45.33333
32 9 3 t2 41 45.33333
33 10 4 t2 35 45.33333
34 11 5 t2 55 45.33333
35 12 6 t2 49 45.33333
36 13 1 t3 55 49.66667
37 14 2 t3 45 49.66667
38 15 3 t3 43 49.66667
39 16 4 t3 40 49.66667
40 17 5 t3 59 49.66667
41 18 6 t3 56 49.66667
42 > (ssw <- sum((xx$value - xx$gr.mean)**2))
43 [1] 715.5
44 > ## sum of squares sub
45 > (sub.xx <- xx %>% group_by(subject) %>% summarise(sub.mean=mean(value)))
46 subject sub.mean
47 1 1 50.00000
48 2 2 43.00000
49 3 3 40.00000
50 4 4 38.00000
51 5 5 55.00000
52 6 6 49.66667
53 > (sssub <- k*sum((sub.xx$sub.mean - mean(xx$value))**2))
54 [1] 658.2778
55 > ## sum of squares error
56 > (sserror <- ssw - sssub)
57 [1] 57.22222
58 > ## ssw
Example
- now it is ease to calculate the meas squares and the F statistic
Example
1 > require(ez)
2 > xx <- as.data.frame(xx)
3 > (an <- ezANOVA(data=xx,dv = value,wid= subject, within = variable))
4 Effect DFn DFd F p p<.05 ges
5 2 variable 2 10 12.53398 0.001885591 * 0.1670008
6 Effect W p p<.05
7 2 variable 0.4335338 0.1879515
8 Effect GGe p[GG] p[GG]<.05 HFe p[HF] p[HF]<.05
9 2 variable 0.6383796 0.008985215 * 0.760165 0.005284458 *
MANOVA
- in general, the framework of multivariate linear models extends quite elegantly to repeated-measure design
- the p responses per individual related to the dependend variables in the model
- the treatment sum of squares and the error sum of squares from the univariate case last week extends to the p x p sum of squares and crossproducts
- test statistics (Wilks lambda, Pillai trace, Hotelling-Lawley trace, Roys maximum root) for testing are based on the non-zero latent roots of
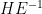
and attempt to capture how large H relative to E
- all these statistics have transformations to F
MANOVA
- the matrix H has the form
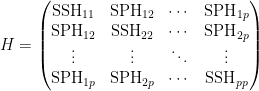
- where e.g.
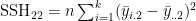
- and e.g.
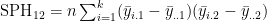
MANOVA
- the matrix E has the form
\vdots & \vdots & \ddots & \vdots \\
- where e.g.
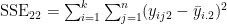
- and e.g.
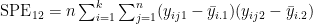
Example Data
- weights of guinea pigs under three levels of vitamin E supplements
- measured six times at the end of week 1, 3, 4, 5, 6, and 7
- contained in guinea.rdata
- Crowder and Hand 1990
Example Data
- load the data print them on the screen
1 > load("guinea.rdata")
2 > head(guinea,10)
3 Dose Animal X1 X3 X4 X5 X6 X7
4 1 0 1 455 460 510 504 436 466
5 2 0 2 467 565 610 596 542 587
6 3 0 3 445 530 580 597 582 619
7 4 0 4 485 542 594 583 611 612
8 5 0 5 480 500 550 528 562 576
9 6 1 6 514 560 565 524 552 597
10 7 1 7 440 480 536 484 567 569
11 8 1 8 495 570 569 585 576 677
12 9 1 9 520 590 610 637 671 702
13 10 1 10 503 555 591 605 649 675
Example Data
- reshape the data for ggplot2
- plot boxplots and means per treatment group \scriptsize
1 > require(reshape2)
2 > dl <- melt(guinea,id.vars = c("Dose","Animal"))
3 > dl$variable <- extract_numeric(dl$variable)
4 > dl$group <- factor(dl$Dose)
5 > head(dl)
6 Dose Animal variable value group
7 1 0 1 1 455 0
8 2 0 2 1 467 0
9 3 0 3 1 445 0
10 4 0 4 1 485 0
11 5 0 5 1 480 0
12 6 1 6 1 514 1
Example Data
<img alt='sesssion2/rma1.png' src='-1' />
Basic multivariate model
- design
\multicolumn{5}{c}{Factor A (Repeated Measures)} \\ \hline |
|
|Subjects |A_1 |A_2 |\cdots |A_p | |
|
|S_{1} |( y_{11} |y_{12} |\cdots |y_{1p} ) |= \mathbf{y^t_{1}} |
|
|S_{2} |( y_{21} |y_{22} |\cdots |y_{2p} ) |= \mathbf{y^t_{2}} |
|
|S_{3} |( y_{31} |y_{32} |\cdots |y_{3p} ) |= \mathbf{y^t_{3}} |
|
|\vdots |\vdots |\vdots | | \vdots o| \vdots |
|
|S_{n} |( y_{n1} |y_{n2} |\cdots |y_{np} ) |= \mathbf{y^t_{n}} |
|
Basic multivariate model
- basic MVLM with an intercept only
- intercepts estimate the means at each instant of time
Basic multivariate model
now test the Null hypothesis #!latex \mu_1=\mu_2=\ldots =\mu_n$
- it is the simplest case of a multivariate test
- extract the sum of squares and products matrix a) for hypothesis
1 > av$SSP
2 X1 X3 X4 X5 X6 X7
3 X1 3545857 3901755 4188127 4133672 4224592 4471095
4 X3 3901755 4293375 4608490 4548570 4648615 4919860
5 X4 4188127 4608490 4946733 4882415 4989803 5280956
6 X5 4133672 4548570 4882415 4818934 4924925 5212293
7 X6 4224592 4648615 4989803 4924925 5033248 5326936
8 X7 4471095 4919860 5280956 5212293 5326936 5637761
- extract the sum of squares and products matrix b) for errors
1 > av$SSPE
2 X1 X3 X4 X5 X6 X7
3 X1 11450.4 10716 5679.20 9326.6 13435.20 14389.80
4 X3 10716.0 19668 13703.00 19888.0 21463.00 25693.00
5 X4 5679.2 13703 13294.93 17357.8 18419.93 19089.73
6 X5 9326.6 19888 17357.80 29166.4 27322.80 29977.20
7 X6 13435.2 21463 18419.93 27322.8 45448.93 42336.73
8 X7 14389.8 25693 19089.73 29977.2 42336.73 47268.93
- size of the (degenerate) ellipse for the intercept term relative to that for Error gives the strength of evidence for the difference between the sample means and the means under
- the H ellipse extends outside the E ellipse (anywhere), this signals that H0 is clearly rejected (for some linear combination of the response variables)
<img alt='sesssion2/rma2.png' src='-1' />
Testing within-S effects
- but up to now there is no information about the within effect structure
- for repeated measurement design we need to specify the intra-subject design
- structure for within-S effects is specified through the arguments idata and idesign
- we need to contruct a time variable for the different measurement times
Test Statistics
- using the Wilks Statistic instead of the default (Pillai)
The 4 Test Statistics
- extract all test statistics
1 > source("functions.r")
2 > get.p.vals.all(av2)
3 Df test stat approx F num Df den Df Pr(>F)
4 time Pillai 1 9.556647e-01 43.1108 5 10 1.899089e-06
5 time Wilks 1 4.433528e-02 43.1108 5 10 1.899089e-06
6 time HL 1 2.155540e+01 43.1108 5 10 1.899089e-06
7 time Roy 1 2.155540e+01 43.1108 5 10 1.899089e-06
The 4 Test Statistics
when #!latex H_0 is true (i.e.
latex error! exitcode was 1 (signal 0), transscript follows: This is pdfTeX, Version 3.14159265-2.6-1.40.19 (TeX Live 2019/dev/Debian) (preloaded format=latex) entering extended mode (./latex_bbe9a80d79364e057925f79143e8266b80751bcb_p.tex LaTeX2e <2018-12-01> (/usr/share/texlive/texmf-dist/tex/latex/base/article.cls Document Class: article 2018/09/03 v1.4i Standard LaTeX document class (/usr/share/texlive/texmf-dist/tex/latex/base/size12.clo)) (/usr/share/texlive/texmf-dist/tex/latex/amsfonts/amssymb.sty (/usr/share/texlive/texmf-dist/tex/latex/amsfonts/amsfonts.sty)) (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsmath.sty For additional information on amsmath, use the `?' option. (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amstext.sty (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsgen.sty)) (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsbsy.sty) (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsopn.sty)) (/usr/share/texlive/texmf-dist/tex/latex/amscls/amsthm.sty) (/usr/share/texlive/texmf-dist/tex/latex/base/inputenc.sty) No file latex_bbe9a80d79364e057925f79143e8266b80751bcb_p.aux. ! You can't use `macro parameter character #' in horizontal mode. l.10 * when {{{# !latex H_0 ! Missing $ inserted. <inserted text> $ l.10 * when {{{#!latex H_ 0 (/usr/share/texlive/texmf-dist/tex/latex/amsfonts/umsa.fd) (/usr/share/texlive/texmf-dist/tex/latex/amsfonts/umsb.fd) ! Missing $ inserted. <inserted text> $ l.11 \end{document} [1] (./latex_bbe9a80d79364e057925f79143e8266b80751bcb_p.aux) ) (\end occurred inside a group at level 3) ### simple group (level 3) entered at line 10 ({) ### simple group (level 2) entered at line 10 ({) ### simple group (level 1) entered at line 10 ({) ### bottom level (see the transcript file for additional information) Output written on latex_bbe9a80d79364e057925f79143e8266b80751bcb_p.dvi (1 page, 268 bytes). Transcript written on latex_bbe9a80d79364e057925f79143e8266b80751bcb_p.log.is false, the four tests have different probabilities of rejectionrelative powers of the four tests depend on the configuration of the mean vectors in the #!latex s$-dimensional space, where {{{#!latex s=\min(p,\nu_H)
indication about this configuration is given by the eigenvalues of #!latex \mathbb{E}^{-1}\mathbb{H}
The 4 Test Statistics
1 > source("functions.r")
2 > get.p.vals.all(av2)
3 Df test stat approx F num Df den Df Pr(>F)
4 time Pillai 1 9.556647e-01 43.1108 5 10 1.899089e-06
5 time Wilks 1 4.433528e-02 43.1108 5 10 1.899089e-06
6 time HL 1 2.155540e+01 43.1108 5 10 1.899089e-06
7 time Roy 1 2.155540e+01 43.1108 5 10 1.899089e-06
The 4 Test Statistics
if one eigenvalue is large and the others are small (collinear mean vectors) Roy is the most powerful (in terms of power: Roy > LH > Wilk > Pillai)
intermediate between collinear and diffuse: Pillai > Wilk > LH > Roy
- Pillai, Wilk, LH are robust against skewness
- Pillai is better in presence of heterogeneity of covariance matrices
- Pillai in most cases the best choice (but performs not well in case of collinearity)
- Wilks not far behind but more sensible in case of severe heterogeneity of covariance matrices
- nowadays software programs calculate all of them and most of the times they lead to the same conclusions, in case of conflicting conclusions examination of eigenvalues and covariance matrices is recommended
The 4 Test Statistics
- calculate the eigenvalues
Add between-S effects
- now add the group term to the model (Dosef)
- design:
Add between-S effects
- design:
\multicolumn{5}{c}{Factor A (Repeated Measures)} \\ \hline |
|
Factor B |Subjects |A_1 |A_2 |\cdots |A_p | |
|
B_1 |S_{11} |( y_{111} |y_{112} |\cdots |y_{11p} ) |= \mathbf{y^t_{11}} |
|
|S_{12} |( y_{121} |y_{122} |\cdots |y_{12p} ) |= \mathbf{y^t_{12}} |
|
|S_{13} |( y_{131} |y_{132} |\cdots |y_{13p} ) |= \mathbf{y^t_{13}} |
|
|\vdots |\vdots |\vdots | | \vdots | \vdots |
|
|S_{1n} |( y_{2n1} |y_{2n2} |\cdots |y_{2np} ) |= \mathbf{y^t_{2n}} |
|
B_2 |S_{21} |( y_{211} |y_{212} |\cdots |y_{21p} ) |= \mathbf{y^t_{21}} |
|
|S_{22} |( y_{221} |y_{222} |\cdots |y_{22p} ) |= \mathbf{y^t_{22}} |
|
|S_{23} |( y_{231} |y_{232} |\cdots |y_{23p} ) |= \mathbf{y^t_{23}} |
|
|\vdots |\vdots |\vdots | | \vdots o| \vdots |
|
|S_{2n} |( y_{2n1} |y_{2n2} |\cdots |y_{2np} ) |= \mathbf{y^t_{2n}} |
|
B_k |S_{k1} |( y_{k11} |y_{k12} |\cdots |y_{k1p} ) |= \mathbf{y^t_{k1}} |
|
|S_{k2} |( y_{k21} |y_{k22} |\cdots |y_{k2p} ) |= \mathbf{y^t_{k2}} |
|
|S_{k3} |( y_{k31} |y_{k32} |\cdots |y_{k3p} ) |= \mathbf{y^t_{k3}} |
|
|\vdots |\vdots |\vdots | | \vdots | \vdots |
|
|S_{kn} |( y_{kn1} |y_{kn2} |\cdots |y_{knp} ) |= \mathbf{y^t_{kn}} |
|
Add between-S effects
1 > (mod2 <- lm(cbind(X1,X3,X4,X5,X6,X7) ~ Dosef,data=guinea))
2 X1 X3 X4 X5 X6 X7
3 Dosef1 28.0 31.6 5.4 5.4 56.4 72.0
4 Dosef2 31.4 15.2 11.0 10.2 41.6 51.2
5 > idata <- data.frame(time)
6 > (av2.mod2 <- Anova(mod2, idata = idata, idesign = ~time))
7 Type II Repeated Measures MANOVA Tests: Pillai test statistic
8 Df test stat approx F num Df den Df Pr(>F)
9 Dosef 2 0.14960 1.1 2 12 0.37821
10 time 1 0.96118 39.6 5 8 1.954e-05 ***
MANOVA Tests
1 > get.p.vals.all(av2.mod2)
2 Df test stat approx F num Df den Df Pr(>F)
3 Dosef Pillai 2 1.496026e-01 1.055525 2 12 3.782088e-01
4 Dosef Wilks 2 8.503974e-01 1.055525 2 12 3.782088e-01
5 Dosef HL 2 1.759208e-01 1.055525 2 12 3.782088e-01
6 Dosef Roy 2 1.759208e-01 1.055525 2 12 3.782088e-01
7 time Pillai 1 9.611815e-01 39.617478 5 8 1.954259e-05
8 time Wilks 1 3.881848e-02 39.617478 5 8 1.954259e-05
9 time HL 1 2.476092e+01 39.617478 5 8 1.954259e-05
10 time Roy 1 2.476092e+01 39.617478 5 8 1.954259e-05
Assumptions MANOVA
- Normal Distribution: The dependent variable should be normally distributed within groups. Overall, the F test is robust to non-normality, if the non-normality is caused by skewness rather than by outliers.
- Linearity: MANOVA assumes that there are linear relationships among all pairs of dependent variables, all pairs of covariates, and all dependent variable-covariate pairs in each cell. Therefore, when the relationship deviates from linearity, the power of the analysis will be compromised.
- Remember that the error variance is computed (SS error) by adding up the sums of squares within each group. If the variances in the two groups are different from each other, then adding the two together is not appropriate, and will not yield an estimate of the common within-group variance. Homoscedasticity can be examined graphically or by means of a number of statistical tests.
- Homogeneity of Variances and Covariances: In multivariate designs, with multiple dependent measures, the homogeneity of variances assumption described earlier also applies. However, since there are multiple dependent variables, it is also required that their intercorrelations (covariances) are homogeneous across the cells of the design. There are various specific tests of this assumption.
- Outliers: Like ANOVA, MANOVA is extremely sensitive to outliers. Outliers may produce either a Type I or Type II error and give no indication as to which type of error is occurring in the analysis. There are several programs available to test for univariate and multivariate outliers.
- Multicollinearity and Singularity: When there is high correlation between dependent variables, one dependent variable becomes a near-linear combination of the other dependent variables. Under such circumstances, it would become statistically redundant and suspect to include both combinations.
Alternative Approach - Mixed Model
the mixed model looks very similar to ANOVA: #!latex Y_{ij} = \mu + \alpha_j + S_i + Error_{ij}$
- the difference is the assumption:
#!latex \mu is a fixed effect: no estimate of variability
#!latex \alpha_j are also fixed effects: no estimate of variability
#!latex S_i are random effects have normal distribution with zero mean
#!latex E_{ij} are normally distributed with mean zero (as usual)
Alternative Approach - Mixed Model
- this also can be written as follows:
#!latex Y_{ij} = [\pi_{0i} + \pi_{2i} \cdot \mathrm{TIME}_{ij}] + [\epsilon_{ij}] (structural part + stochastic part)
#!latex Y_{ij} is the value of subject #!latex i at time #!latex j as a linear function of #!latex \mathrm{TIME}
#!latex \pi_{0i} and #!latex \pi_{2i} are individual parameters that characterize its shape for the
latex error! exitcode was 1 (signal 0), transscript follows: This is pdfTeX, Version 3.14159265-2.6-1.40.19 (TeX Live 2019/dev/Debian) (preloaded format=latex) entering extended mode (./latex_00683f2d325b5e274d8b6f04699fec10646ec9c7_p.tex LaTeX2e <2018-12-01> (/usr/share/texlive/texmf-dist/tex/latex/base/article.cls Document Class: article 2018/09/03 v1.4i Standard LaTeX document class (/usr/share/texlive/texmf-dist/tex/latex/base/size12.clo)) (/usr/share/texlive/texmf-dist/tex/latex/amsfonts/amssymb.sty (/usr/share/texlive/texmf-dist/tex/latex/amsfonts/amsfonts.sty)) (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsmath.sty For additional information on amsmath, use the `?' option. (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amstext.sty (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsgen.sty)) (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsbsy.sty) (/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsopn.sty)) (/usr/share/texlive/texmf-dist/tex/latex/amscls/amsthm.sty) (/usr/share/texlive/texmf-dist/tex/latex/base/inputenc.sty) No file latex_00683f2d325b5e274d8b6f04699fec10646ec9c7_p.aux. ! You can't use `macro parameter character #' in horizontal mode. l.10 * {{{# !latex \gamma_{00 ! Missing $ inserted. <inserted text> $ l.10 * {{{#!latex \gamma _{00 (/usr/share/texlive/texmf-dist/tex/latex/amsfonts/umsa.fd) (/usr/share/texlive/texmf-dist/tex/latex/amsfonts/umsb.fd) ! Missing $ inserted. <inserted text> $ l.11 \end{document} ! Missing } inserted. <inserted text> } l.11 \end{document} [1] (./latex_00683f2d325b5e274d8b6f04699fec10646ec9c7_p.aux) ) (\end occurred inside a group at level 3) ### simple group (level 3) entered at line 10 ({) ### simple group (level 2) entered at line 10 ({) ### simple group (level 1) entered at line 10 ({) ### bottom level (see the transcript file for additional information) Output written on latex_00683f2d325b5e274d8b6f04699fec10646ec9c7_p.dvi (1 page, 308 bytes). Transcript written on latex_00683f2d325b5e274d8b6f04699fec10646ec9c7_p.log.} and #!latex \gamma_{10} the level-2 intercepts, represent the population average initial status and rate of change where #!latex \mathrm{X}=0
the level-2 slopes #!latex \gamma_{01} and #!latex \gamma_{11} represent the treatment effect #!latex \mathrm{X} on the intercept and the slope respectively
#!latex \xi_{0i} and #!latex \xi_{1i}, the level-2 residuals represent those portions of initial status or rate of change that are unexplained at level-2, they represent deviations of the individual change trajectories around their respective group average trends
#!latex \xi_{0i} and #!latex \xi_{1i} are assumed to be independently draw from a bivariate normal distribution with mean 0 and variances #!latex \sigma_0^2 and #!latex \sigma_1^2
Alternative Approach - Mixed Model
Alternative Approach - Mixed Model
Alternative Approach - Mixed Model
- extract fixed effects
Alternative Approach - Mixed Model
- post hoc test
1 > summary(glht(Lme.mod, linfct=mcp(group="Tukey")))
2 Multiple Comparisons of Means: Tukey Contrasts
3 Linear Hypotheses:
4 Estimate Std. Error z value Pr(>|z|)
5 1 - 0 == 0 28.00 27.69 1.011 0.570
6 2 - 0 == 0 31.40 27.69 1.134 0.493
7 2 - 1 == 0 3.40 27.69 0.123 0.992
8 In mcp2matrix(model, linfct = linfct) :
9 covariate interactions found -- default contrast might be inappropriate
Mixed Models
- average the interaction term
1 > ## consider interaction terms
2 > summary(glht(Lme.mod, linfct=mcp(group="Tukey",interaction_average = TRUE)))
3 Multiple Comparisons of Means: Tukey Contrasts
4 Linear Hypotheses:
5 Estimate Std. Error z value Pr(>|z|)
6 1 - 0 == 0 33.133 24.202 1.369 0.357
7 2 - 0 == 0 26.767 24.202 1.106 0.510
8 2 - 1 == 0 -6.367 24.202 -0.263 0.963
Mixed Models
1 > Lme.mod <- lmer(value ~ group + variable + (1 | Animal), data = dl)
2 > anova(Lme.mod)
3 Analysis of Variance Table
4 Df Sum Sq Mean Sq F value
5 group 2 1276 638.1 1.0555
6 variable 5 142555 28510.9 47.1641
7 > summary(glht(Lme.mod, linfct=mcp(group="Tukey")))
8 Simultaneous Tests for General Linear Hypotheses
9 Multiple Comparisons of Means: Tukey Contrasts
10 Linear Hypotheses:
11 Estimate Std. Error z value Pr(>|z|)
12 1 - 0 == 0 33.133 24.202 1.369 0.357
13 2 - 0 == 0 26.767 24.202 1.106 0.510
14 2 - 1 == 0 -6.367 24.202 -0.263 0.963
Exercises
* try to set up an unconditional means model using lme or lmer from the respective package. * Extract the coefficients using coef() and interpret the results. * add time as fixed effect. * add time as random effect. Again: extract the coefficients, interpret. * add the group variable as factor, do an posthoc test
Solutions
* try to set up an unconditional means model using lme or lmer from the respective package.\scriptsize
1 > mod.0 <- lmer(value ~ 1 + (1 | Animal ), data = dl)
* Extract the coefficients using coef() and interpret the results.\scriptsize
* add time as fixed effect.\scriptsize
1 > mod.1 <- lmer(value ~ variable + (1 | Animal ), data = dl)
2 > coef(mod.1)
3 1 394.8259 19.40286
4 2 476.9201 19.40286
5 3 474.7759 19.40286
6 4 486.1098 19.40286
7 5 450.7296 19.40286
8 6 468.4963 19.40286
9 7 432.3503 19.40286
10 8 493.0020 19.40286
11 9 532.5175 19.40286
12 10 509.2371 19.40286
13 11 512.9129 19.40286
14 12 476.0012 19.40286
15 13 465.4331 19.40286
16 14 508.4713 19.40286
17 15 443.5311 19.40286
* add time as random effect. Again: extract the coefficients, interpret.\scriptsize
1 > mod.2 <- lmer(value ~ variable + (1 + variable | Animal ), data = dl)
2 > coef(mod.2)
3 1 452.6571 4.989677
4 2 475.9532 19.079395
5 3 474.2833 20.463988
6 4 478.1603 21.318140
7 5 468.0379 15.382203
8 6 473.9412 17.008690
9 7 462.0846 13.445867
10 8 480.0258 22.650268
11 9 491.3526 29.244908
12 10 484.4619 25.718884
13 11 485.6489 26.112158
14 12 475.6310 19.023129
15 13 472.0510 18.168807
16 14 485.1363 24.115063
17 15 465.8891 14.321680
18 [1] "coef.mer"
* add the group variable as factor, do an posthoc test\scriptsize
1 > mod.3 <- lmer(value ~ variable + group + (1 + variable | Animal ), data = dl)
2 > coef(mod.3)
3 1 448.2977 5.81501 10.60605 14.33476
4 2 472.9694 19.59789 10.60605 14.33476
5 3 465.2712 22.15993 10.60605 14.33476
6 4 473.0687 22.24370 10.60605 14.33476
7 5 461.8873 16.53096 10.60605 14.33476
8 6 469.0268 15.91834 10.60605 14.33476
9 7 447.5350 14.25275 10.60605 14.33476
10 8 470.6132 22.42630 10.60605 14.33476
11 9 483.2428 28.74823 10.60605 14.33476
12 10 474.3565 25.62254 10.60605 14.33476
13 11 475.0372 25.41345 10.60605 14.33476
14 12 466.9784 17.95933 10.60605 14.33476
15 13 459.9408 17.78472 10.60605 14.33476
16 14 478.6720 22.60904 10.60605 14.33476
17 15 453.7132 13.96066 10.60605 14.33476
18 [1] "coef.mer"
19 > summary(glht(mod.3, linfct=mcp(group="Tukey")))
20 Multiple Comparisons of Means: Tukey Contrasts
21 data = dl)
22 Linear Hypotheses:
23 Estimate Std. Error z value Pr(>|z|)
24 1 - 0 == 0 10.606 16.669 0.636 0.800
25 2 - 0 == 0 14.335 16.669 0.860 0.666
26 2 - 1 == 0 3.729 16.669 0.224 0.973
