|
Größe: 17701
Kommentar:
|
Größe: 17855
Kommentar:
|
| Gelöschter Text ist auf diese Art markiert. | Hinzugefügter Text ist auf diese Art markiert. |
| Zeile 44: | Zeile 44: |
| ||nu ||degrees of freedom| | ||nu ||degrees of freedom|| |
| Zeile 55: | Zeile 55: |
| <img alt='sesssion2/twosided.pdf}' src='-1' /> <img alt='sesssion2/greater.pdf}' src='-1' /> <img alt='sesssion2/less.pdf}' src='-1' /> == Alternatives == The p-value is the probability of the sample estimate (of the respective estimator) under the null. |
[[attachment:twosided.pdf|{{attachment:twosided.pdf||width=800,height=400}}]] [[attachment:greater.pdf|{{attachment:greater.pdf||width=800,height=400}}]] [[attachment:less.pdf|{{attachment:less.pdf||width=800,height=400}}]] The p-value is the probability of the sample estimate (of the respective estimator) under the null. The p-value is NOT the probability that the null is true. |
| Zeile 62: | Zeile 65: |
| To investigate the significance of the difference between an assumed population mean {{{#!latex \mu_0}}} and a sample mean {{{#!latex \bar{x}}}}. * It is necessary that the population variance {{{#!latex \sigma^2}}} is known. * The test is accurate if the population is normally distributed. If the population is not normal, the test will still give an approximate guide. |
To investigate the significance of the difference between an assumed population mean {{{#!latex $\mu_0$}}} and a sample mean {{{#!latex $\bar{x}$ }}}. * It is necessary that the population variance {{{#!latex \sigma^2}}} is known. * The test is accurate if the population is normally distributed. If the population is not normal, the test will still give an approximate guide. |
Classical Tests
Exercises
- load the data (file: session4data.rdata)
- make a new summary data frame (per subject and time) containing:
- the number of trials
- the number correct trials (absolute and relative)
- the mean TTime and the standard deviation of TTime
- the respective standard error of the mean
- keep the information about Sex and Age\_PRETEST
- make a plot with time on the x-axis and TTime on the y-axis showing the means and the 95\% confidence intervals (geom\_pointrange())
- add the number of trials and the percentage of correct ones using geom\_text()
Exercises - Solutions
- load the data (file: session4data.rdata)
- make a new summary data frame (per subject and time) containing:
1 > sumdf <- data %>%
2 + group_by(Subject,Sex,Age_PRETEST,testid) %>%
3 + summarise(count=n(),
4 + n.corr = sum(Stim.Type=="hit"),
5 + perc.corr = n.corr/count,
6 + mean.ttime = mean(TTime),
7 + sd.ttime = sd(TTime),
8 + se.ttime = sd.ttime/sqrt(count))
9 > head(sumdf)
10 Subject Sex Age_PRETEST testid count n.corr perc.corr mean.ttime
11 1 1 f 3.11 test1 95 63 0.6631579 8621.674
12 2 1 f 3.11 1 60 32 0.5333333 9256.367
13 3 1 f 3.11 2 59 32 0.5423729 9704.712
14 4 1 f 3.11 3 60 38 0.6333333 14189.550
15 5 1 f 3.11 4 59 31 0.5254237 13049.831
16 6 1 f 3.11 5 59 33 0.5593220 14673.525
17 Variables not shown: sd.ttime se.ttime (dbl)
four possible situations
|
Situation |
||
|
H_0 is true |
H_0 is false |
|
Conclusion |
H_0 is not rejected |
Correct decision |
Type II error |
H_0 is rejected |
Type I error |
Correct decision |
|
Common symbols
n |
number of observations (sample size) |
K |
number of samples (each having n elements) |
alpha |
level of significance |
nu |
degrees of freedom |
mu |
population mean |
xbar |
sample mean |
sigma |
standard deviation (population) |
s |
standard deviation (sample) |
rho |
population correlation coefficient |
r |
sample correlation coefficient |
Z |
standard normal deviate |
Alternatives
The p-value is the probability of the sample estimate (of the respective estimator) under the null. The p-value is NOT the probability that the null is true.
Z-test for a population mean
The z-test is a something like a t-test (it is like you would know almost everything about the perfect conditions. It uses the normal distribution as test statistic and is therefore a good example. To investigate the significance of the difference between an assumed population mean
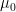
and a sample mean
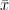
.
It is necessary that the population variance #!latex \sigma^2 is known.
- The test is accurate if the population is normally distributed. If the population is not normal, the test will still give an approximate guide.
Z-test for a population mean
* Write a function which takes a vector, the population standard deviation and the population mean as arguments and which gives the Z score as result.
- name the function ztest or my.z.test - not z.test because z.test is already used
- set a default value for the population mean
* add a line to your function that allows you to process numeric vectors containing missing values! * the function pnorm(Z) gives the probability of
latex error! exitcode was 1 (signal 0), transscript follows:
This is pdfTeX, Version 3.14159265-2.6-1.40.19 (TeX Live 2019/dev/Debian) (preloaded format=latex)
entering extended mode
(./latex_3804b1112b760080c8c10d0951a569ca7c0b3197_p.tex
LaTeX2e <2018-12-01>
(/usr/share/texlive/texmf-dist/tex/latex/base/article.cls
Document Class: article 2018/09/03 v1.4i Standard LaTeX document class
(/usr/share/texlive/texmf-dist/tex/latex/base/size12.clo))
(/usr/share/texlive/texmf-dist/tex/latex/amsfonts/amssymb.sty
(/usr/share/texlive/texmf-dist/tex/latex/amsfonts/amsfonts.sty))
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsmath.sty
For additional information on amsmath, use the `?' option.
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amstext.sty
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsgen.sty))
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsbsy.sty)
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsopn.sty))
(/usr/share/texlive/texmf-dist/tex/latex/amscls/amsthm.sty)
(/usr/share/texlive/texmf-dist/tex/latex/base/inputenc.sty)
No file latex_3804b1112b760080c8c10d0951a569ca7c0b3197_p.aux.
! You can't use `macro parameter character #' in horizontal mode.
l.14 {{{#
!highlight r
[1] (./latex_3804b1112b760080c8c10d0951a569ca7c0b3197_p.aux) )
(\end occurred inside a group at level 3)
### simple group (level 3) entered at line 14 ({)
### simple group (level 2) entered at line 14 ({)
### simple group (level 1) entered at line 14 ({)
### bottom level
(see the transcript file for additional information)
Output written on latex_3804b1112b760080c8c10d0951a569ca7c0b3197_p.dvi (1 page,
944 bytes).
Transcript written on latex_3804b1112b760080c8c10d0951a569ca7c0b3197_p.log.
Z-test for a population mean
Add a line to your function that allows you to also process numeric vectors containing missing values!
Z-test for a population mean
The function pnorm(Z) gives the probability of
latex error! exitcode was 1 (signal 0), transscript follows:
This is pdfTeX, Version 3.14159265-2.6-1.40.19 (TeX Live 2019/dev/Debian) (preloaded format=latex)
entering extended mode
(./latex_0d342107de9429eef76138cc515ca3fe22cb439b_p.tex
LaTeX2e <2018-12-01>
(/usr/share/texlive/texmf-dist/tex/latex/base/article.cls
Document Class: article 2018/09/03 v1.4i Standard LaTeX document class
(/usr/share/texlive/texmf-dist/tex/latex/base/size12.clo))
(/usr/share/texlive/texmf-dist/tex/latex/amsfonts/amssymb.sty
(/usr/share/texlive/texmf-dist/tex/latex/amsfonts/amsfonts.sty))
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsmath.sty
For additional information on amsmath, use the `?' option.
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amstext.sty
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsgen.sty))
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsbsy.sty)
(/usr/share/texlive/texmf-dist/tex/latex/amsmath/amsopn.sty))
(/usr/share/texlive/texmf-dist/tex/latex/amscls/amsthm.sty)
(/usr/share/texlive/texmf-dist/tex/latex/base/inputenc.sty)
No file latex_0d342107de9429eef76138cc515ca3fe22cb439b_p.aux.
! You can't use `macro parameter character #' in vertical mode.
l.10 {{{#
!highlight r
Overfull \hbox (7.40698pt too wide) in paragraph at lines 10--20
[]\OT1/cmr/m/n/12 !highlight r > ztest <- func-tion(x,x.sd,mu=0) + x <- x[!is.n
a(x)] + if(length(x)
[1] (./latex_0d342107de9429eef76138cc515ca3fe22cb439b_p.aux) )
(\end occurred inside a group at level 3)
### simple group (level 3) entered at line 10 ({)
### simple group (level 2) entered at line 10 ({)
### simple group (level 1) entered at line 10 ({)
### bottom level
(see the transcript file for additional information)
Output written on latex_0d342107de9429eef76138cc515ca3fe22cb439b_p.dvi (1 page,
480 bytes).
Transcript written on latex_0d342107de9429eef76138cc515ca3fe22cb439b_p.log.
Z-test for a population mean
Now let the result be a named vector containing the estimated difference, Z, p and the n.
1 > ztest <- function(x,x.sd,mu=0){
2 + x <- x[!is.na(x)]
3 + if(length(x) < 3) stop("too few values in x")
4 + est.diff <- mean(x)-mu
5 + z <- sqrt(length(x)) * (est.diff)/x.sd
6 + round(c(diff=est.diff,Z=z,pval=2*pnorm(-abs(z)),n=length(x)),4)
7 + }
8 > set.seed(1)
9 > ztest(rnorm(100),x.sd = 1)
10 diff Z pval n
Z-test for a population mean
* Z-test for two population means (variances known and equal) * Z-test for two population means (variances known and unequal) To investigate the statistical significance of the difference between an assumed population mean #!latex \mu_0 and a sample mean #!latex \bar{x}. There is a function z.test() in the BSDA package. * It is necessary that the population variance #!latex \sigma^2 is known. * The test is accurate if the population is normally distributed. If the population is not normal, the test will still give an approximate guide.
Simulation Exercises
* Now sample 100 values from a Normal distribution with mean 10 and standard deviation 2 and use a z-test to compare it against the population mean 10. What is the p-value? * Now do the sampling and the testing 1000 times, what would be the number of statistically significant results? Use replicate() (which is a wrapper of tapply()) or a for() loop! Record at least the p-values and the estimated differences! Use table() to count the p-vals below 0.05. What type of error do you associate with it? What is the smallest absolute difference with a p-value below 0.05? * Repeat the simulation above, change the sample size to 1000 in each of the 1000 samples! How many p-values below 0.05? What is now the smallest absolute difference with a p-value below 0.05?
Simulation Exercises -- Solutions
- Now sample 100 values from a Normal distribution with mean 10 and standard deviation 2 and use a z-test to compare it against the population mean 10. What is the p-value? What the estimated difference?
Simulation Exercises -- Solutions
- Now do the sampling and the testing 1000 times, what would be the number of statistically significant results? Use replicate() (which is a wrapper of tapply()) or a for() loop. Record at least the p-values and the estimated differences! Transform the result into a data frame.
using replicate()\footnotesize
1 > res <- replicate(1000, ztest(rnorm(100,mean=10,sd=2),x.sd=2,mu=10))
2 > res <- as.data.frame(t(res))
3 > head(res)
4 diff Z pval n
5 1 -0.2834 -1.4170 0.1565 100
6 2 0.2540 1.2698 0.2042 100
7 3 -0.1915 -0.9576 0.3383 100
8 4 0.1462 0.7312 0.4646 100
9 5 0.1122 0.5612 0.5747 100
10 6 -0.0141 -0.0706 0.9437 100
Simulation Exercises -- Solutions
- Now do the sampling and the testing 1000 times, what would be the number of statistically significant results? Use replicate() (which is a wrapper of tapply()) or a for() loop. Record at least the p-values and the estimated differences! Transform the result into a data frame.
using replicate() II\footnotesize
Simulation Exercises -- Solutions
- Now do the sampling and the testing 1000 times, what would be the number of statistically significant results? Use replicate() (which is a wrapper of tapply()) or a for() loop. Record at least the p-values and the estimated differences! Transform the result into a data frame.
using for() \scriptsize
1 > res <- matrix(numeric(2000),ncol=2)
2 > for(i in seq.int(1000)){
3 + res[i,] <- ztest(rnorm(100,mean=10,sd=2),x.sd=2,mu=10)[c("pval","diff")] }
4 > res <- as.data.frame(res)
5 > names(res) <- c("pval","diff")
6 > head(res)
7 pval diff
8 1 0.0591 -0.3775
9 2 0.2466 0.2317
10 3 0.6368 0.0944
11 4 0.5538 -0.1184
12 5 0.9897 -0.0026
13 6 0.7748 0.0572
Simulation Exercises -- Solutions
- Use table() to count the p-vals below 0.05. What type of error do you associate with it? What is the smallest absolute difference with a p-value below 0.05?
Simulation Exercises -- Solutions
- Repeat the simulation above, change the sample size to 1000 in each of the 1000 samples! How many p-values below 0.05? What is now the smallest absolute difference with a p-value below 0.05?
1 > res2 <- replicate(1000, ztest(rnorm(1000,mean=10,sd=2),
2 + x.sd=2,mu=10))
3 > res2 <- as.data.frame(t(res2))
4 > head(res2)
5 diff Z pval n
6 1 -0.0731 -1.1559 0.2477 1000
7 2 0.0018 0.0292 0.9767 1000
8 3 0.0072 0.1144 0.9089 1000
9 4 -0.1145 -1.8100 0.0703 1000
10 5 -0.1719 -2.7183 0.0066 1000
11 6 0.0880 1.3916 0.1640 1000
Simulation Exercises -- Solutions
- Repeat the simulation above, change the sample size to 1000 in each of the 1000 samples! How many p-values below 0.05? What is now the smallest absolute difference with a p-value below 0.05?
Simulation Exercises Part II
* Concatenate the both resulting data frames from above using rbind() * Plot the distributions of the pvals and the difference per sample size. Use ggplot2 with an appropriate geom (density/histogram) * What is the message?
Simulation Exercises -- Solutions
- Concatenate the both resulting data frames from above using rbind()
- Plot the distributions of the pvals and the difference per sample size. Use ggplot2 with an appropriate geom (density/histogram)
Simulation Exercises -- Solutions
<img alt='sesssion2/hist.png' src='-1' />
Simulation Exercises -- Solutions
- Plot the distributions of the pvals and the difference per sample size. Use ggplot2 with an appropriate geom (density/histogram)
Simulation Exercises -- Solutions
<img alt='sesssion2/dens.png' src='-1' />
Simulation Exercises -- Solutions
<img alt='sesssion2/point.png' src='-1' />
Simulation Exercises -- Solutions
<img alt='sesssion2/dens2d.png' src='-1' />
t-tests
A t-test is any statistical hypothesis test in which the test statistic follows a \emph{Student's t distribution} if the null hypothesis is supported.
- \emph{one sample t-test}: test a sample mean against a population mean
One Sample t-test
Two Sample t-tests
There are two ways to perform a two sample t-test in R:
- given two vectors x and y containing the measurement values from the respective groups t.test(x,y)
given one vector x containing all the measurement values and one vector g containing the group membership #!latex t.test(x \sim g) (read: x dependend on g)
Two Sample t-tests: two vector syntax
1 > set.seed(1)
2 > x <- rnorm(12)
3 > y <- rnorm(12)
4 > g <- sample(c("A","B"),12,replace = T)
5 > t.test(x,y)
6 t = 0.5939, df = 20.012, p-value = 0.5592
7 alternative hypothesis: true difference in means is not equal to 0
8 95 percent confidence interval:
9 sample estimates:
10 mean of x mean of y
Two Sample t-tests: formula syntax
Welch/Satterthwaite vs. Student
- if not stated otherwise t.test() will not assume that the variances in the both groups are equal
- if one knows that both populations have the same variance set the var.equal argument to TRUE to perform a student's t-test
Student's t-test
t-test
- the t-test, especially the Welch test is appropriate whenever the values are normally distributed
it is also recommended for group sizes #!latex \geq 30 (robust against deviation from normality)
Exercises
* use a t-test to compare TTime according to Stim.Type, visualize it. What is the problem? * now do the same for Subject 1 on pre and post test (use filter() or indexing to get the resp. subsets) * use the following code to do the test on every subset Subject and testid, try to figure what is happening in each step:\tiny
* make plots to visualize the results. * how many tests have a statistically significant result? How many did you expect? Is there a tendency? What could be the next step?
Exercises - Solutions
- use a t-test to compare TTime according to Stim.Type, visualize it. What is the problem?
1 > t.test(data$TTime ~ data$Stim.Type)
2 t = -6.3567, df = 9541.891, p-value = 2.156e-10
3 alternative hypothesis: true difference in means is not equal to 0
4 95 percent confidence interval:
5 sample estimates:
6 mean in group hit mean in group incorrect
7 > ggplot(data,aes(x=Stim.Type,y=TTime)) +
8 + geom_boxplot()
Exercises - Solutions
- now do the same for Subject 1 on pre and post test (use filter() or indexing to get the resp. subsets)
1 > t.test(data$TTime[data$Subject==1 & data$testid=="test1"] ~
2 + data$Stim.Type[data$Subject==1 & data$testid=="test1"])
3 t = -0.5846, df = 44.183, p-value = 0.5618
4 alternative hypothesis: true difference in means is not equal to 0
5 95 percent confidence interval:
6 sample estimates:
7 mean in group hit mean in group incorrect
8 > t.test(data$TTime[data$Subject==1 & data$testid=="test2"] ~
9 + data$Stim.Type[data$Subject==1 & data$testid=="test2"])
10 t = -1.7694, df = 47.022, p-value = 0.08332
11 alternative hypothesis: true difference in means is not equal to 0
12 95 percent confidence interval:
13 sample estimates:
14 mean in group hit mean in group incorrect
Exercises - Solutions
- make plots to visualize the results
Exercises - Solutions
- how many tests have an statistically significant result? How many did you expect?
Exercises - Solutions
- What could be the next step?
