Introduction
Remember: Implicit Loops
A common application of loops is to apply a function to each element of a set of values and collect the results in a single structure. In R this is done by the functions:
- lapply() - works on elements of a list
- sapply() - same as lapply but simplify results
- apply() - works on rows or colums of a matrix or a data frame (or more general on arrays)
- tapply() - works on groups defined by an index
Exercises
Given the two R object m and l below use * use lapply() to get the class and the length of each element of l (two steps) * apply() to get the maximum of each column in m
read one file
read several files
- we used dir() (with the arguments pattern, recursive, full.path) to get a list of file names we wanted to read in
- we learned about lapply() which takes a list l and a function f to perform the function f on every element of the list l
- so now we combine what we learned to read all files at once
get files names
1 > files <- dir("../session2/data",full.names = T,
2 + recursive = T,pattern = "[0-9]{3}\\.txt$")
3 > files
4 [1] "../session2/data/posttest/post_001.txt"
5 [2] "../session2/data/posttest/post_002.txt"
6 [3] "../session2/data/posttest/post_003.txt"
7 [4] "../session2/data/posttest/post_004.txt"
8 [5] "../session2/data/posttest/post_005.txt"
9 [6] "../session2/data/posttest/post_006.txt"
10 [7] "../session2/data/posttest/post_007.txt"
read in files
- source the file containing our function read.file()
- use lapply() to use read.file() on every entry of the list of file names
1 > source("function.r")
2 > df.list <- lapply(files,read.file,skip=0)
3 [1] "read ../session2/data/posttest/post_001.txt"
4 [1] "read ../session2/data/posttest/post_002.txt"
5 [1] "read ../session2/data/posttest/post_003.txt"
6 [1] "read ../session2/data/posttest/post_004.txt"
7 [1] "read ../session2/data/posttest/post_005.txt"
8 [1] "read ../session2/data/posttest/post_006.txt"
9 [1] "read ../session2/data/posttest/post_007.txt"
10 [1] "read ../session2/data/posttest/post_008.txt"
Result
- what we get is a list df.list containing the results: every element of the list is a data frame if read.file() read in successfully the respective file
- so our variable files contains 195 file names
- so df.list contains 195 elements
- we can check the class of each of these results again with sapply()
Remember: Combining Data Frames
We learned about three basis functions to combine data frame
- rbind() - combine two data frames row wise
- cbind() - combine two data frames column wise
- merge() - combine two data with respect two one or more identifying columns
- all of them are binary function
- so you can not put more than two data frame into it
- using only these function it would be a tedious and boring work to combine 192 data frames
Reduce()
- is also a higher order function (functional)
- Reduce() uses a binary function (like rbind() or merge()) to combine successively the elements of a given list
- it can be used if you have not only two but many data frames
Example
- first we make up 4 artifical data frames
1 > (d1 <- data.frame(id=LETTERS[c(1,2,3)],day1=sample(10,3)))
2 id day1
3 1 A 3
4 2 B 1
5 3 C 7
6 > (d2 <- data.frame(id=LETTERS[c(1,3,5,6)],day2=sample(10,4)))
7 id day2
8 1 A 8
9 2 C 2
10 3 E 5
11 4 F 3
12 > (d3 <- data.frame(id=LETTERS[c(2,4:6)],day3=sample(10,4)))
13 id day3
14 1 B 8
15 2 D 3
16 3 E 4
17 4 F 10
18 > (d4 <- data.frame(id=LETTERS[c(1:5)],day4=sample(10,5)))
19 id day4
20 1 A 2
21 2 B 7
22 3 C 8
23 4 D 9
24 5 E 1
- now we use Reduce() in combination with merge()
- and what we get is an empty data frame
- well this isn't exactly what we wanted, so why?
- it is because the default behavior of merge() is set all=F, so we get only complete lines which is in this case - none
- so we have to define a wrapper function which only change this argument to all=T
- set all to TRUE
- which is exactly what we want
- a second example in combination with rbind()
- which is exactly what we want
Reduce() Exercise
- the list ml contains three vectors
- use lapply() to get the class of each of them
- then use Reduce() and combination with c() to coerce them into one vector. Of which class is the resulting vector?
Reduce() Exercise
- use lapply() to get the class of each of them
- then use Reduce() and combination with c() to coerce them into one vector. Of which class is the resulting vector?
Combine all data frames
- We used lapply() and our function read.file() to read all files in files
- and we got back a list df.list containing 192 data frames
Combine all data frames - exercise
- now use what we learned about Reduce{} and combining data frames using rbind() to combine these 192 data frames.
Combine all data frames - exercise
1 > data <- Reduce(rbind,df.list)
2 > nrow(data)
3 [1] 12704
4 > table(data$Subject)
5 001_test2 002_test2 003_test2 004_test2 005_test2 006_test2 007_test2 008_test2
6 93 91 96 93 95 95 93 96
7 009_test2 010_test2 011_test2 012_test2 013_test2 014_test2 015_test2 016_test2
8 92 94 95 96 96 95 96 94
9 017_test2 018_test2 019_test2 020_test2 001_test1 002_test1 003_test1 004_test1
10 95 94 96 95 95 95 96 94
11 005_test1 006_test1 007_test1 008_test1 009_test1 010_test1 011_test1 012_test1
12 96 95 94 90 96 95 91 96
13 013_test1 014_test1 015_test1 016_test1 017_test1 018_test1 019_test1 020_test1
14 95 96 95 91 96 96 96 96
15 001_1 002_1 003_1 004_1 005_1 006_1 CHGU_1 008_1a
16 60 59 60 54 60 59 60 60
17 009_1 010_1 RMK_1 013_1 014_1 015_1 016_1 IJ2K_1
18 60 60 59 59 60 58 59 58
19 018_1 019_1 020_1 001_2 002_2 003_2 004_2 005_2
20 60 59 60 59 59 57 58 57
21 006_2 007_2 008_2 009_2 010_2 011_2 012_2 013_2
22 58 58 54 58 58 59 59 56
23 014_2 015_2 016_2 017_2 018_2 019_2 020_2 001_3
The Function no 2
- so it is recommended to build again a function out of this
1 > read.files <- function(filesdir,skip=3,recursive=F,pattern="."){
2 + files <- dir(filesdir,
3 + full.names = T,
4 + recursive = recursive,
5 + pattern = pattern)
6 + Reduce(rbind,lapply(files,read.file,skip=skip))}
7 > data <- read.files("data",recursive = T,skip=0,pattern = "\\.txt$")
8 [1] "read data/posttest/post_001.txt"
9 [1] "read data/posttest/post_002.txt"
10 [1] "read data/posttest/post_003.txt"
11 [1] "read data/posttest/post_004.txt"
12 [1] "read data/posttest/post_005.txt"
The Function no 2
- by changing the pattern (passed through to dir() we can limit the read in files to specific time or person\tiny
1 > sub1 <- read.files("../session2/data",
2 + skip = 0, recursive = T,pattern="\\002\\.txt$")
3 [1] "read ../session2/data/posttest/post_002.txt"
4 [1] "read ../session2/data/pretest/pre_002.txt"
5 [1] "read ../session2/data/training_1/train_002.txt"
6 [1] "read ../session2/data/training_2/train_002.txt"
7 [1] "read ../session2/data/training_3/train_002.txt"
8 > test <- read.files("../session2/data",
9 + skip = 0, recursive = T,pattern="p[ro].+\\.txt$")
10 [1] "read ../session2/data/posttest/post_001.txt"
11 [1] "read ../session2/data/posttest/post_002.txt"
12 [1] "read ../session2/data/pretest/pre_001.txt"
13 [1] "read ../session2/data/pretest/pre_002.txt"
The Subject column
- table the Subject column again. What is the problem?
The Subject column
1 > table(data$Subject)
2 001_test2 002_test2 003_test2 004_test2 005_test2 006_test2 007_test2 008_test2
3 93 91 96 93 95 95 93 96
4 009_test2 010_test2 011_test2 012_test2 013_test2 014_test2 015_test2 016_test2
5 92 94 95 96 96 95 96 94
6 017_test2 018_test2 019_test2 020_test2 001_test1 002_test1 003_test1 004_test1
7 95 94 96 95 95 95 96 94
- subject and time coded in one variable
The Subject column
- we create two new variables using the str_split() function (stringr package)
- because str_split() has a list containing a vector as result we have to use it in combination with sapply()
- then correct some of the person ids
The Subject column
- a alternative is using again regular expressions using the str_replace() function (again stringr package)
- str_replace() takes three arguments: the string, the pattern to be replaced and the replacement
The Subject column
- now table the personid column
- what is left to do?
The Subject column
The Subject column Exercises
- there are some more wrong person ids: RMK - 011, IJ2K - 017, GA3K - 004, Kj6K - 006. Correct them!
The Subject column Exercises
Merging
* now read in the file subjectsdemographics.txt using the appropriate command * join the demographics with our data data frame (there is a little problem left - compare the persid and Subject columns)
The Subject column Exercises
1 > persdat <- read.table("data/subjectdemographics.txt",
2 + sep="\t",
3 + header=T)
4 > persdat$Subject
5 [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
6 > unique(data$persid)
7 [1] "001" "002" "003" "004" "005" "006" "007" "008" "009" "010" "011" "012"
8 [13] "013" "014" "015" "016" "017" "018" "019" "020"
9 > data$persid <- as.numeric(data$persid)
10 > unique(data$persid)
11 [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
12 > data <- merge(persdat,data,by.x = "Subject",by.y = "persid",all=T)
13 > head(data)
14 > summary(data)
15 Subject Sex Age_PRETEST Trial Event.Type
16 1st Qu.: 5.00 m:5046 1st Qu.:3.110 1st Qu.:112.0 Response:12704
17 Median :11.00 Median :4.400 Median :222.0 Sound : 0
18 Mean :10.53 Mean :4.154 Mean :223.1 Pause : 0
19 3rd Qu.:16.00 3rd Qu.:4.600 3rd Qu.:332.0 Resume : 0
Summary Graphics
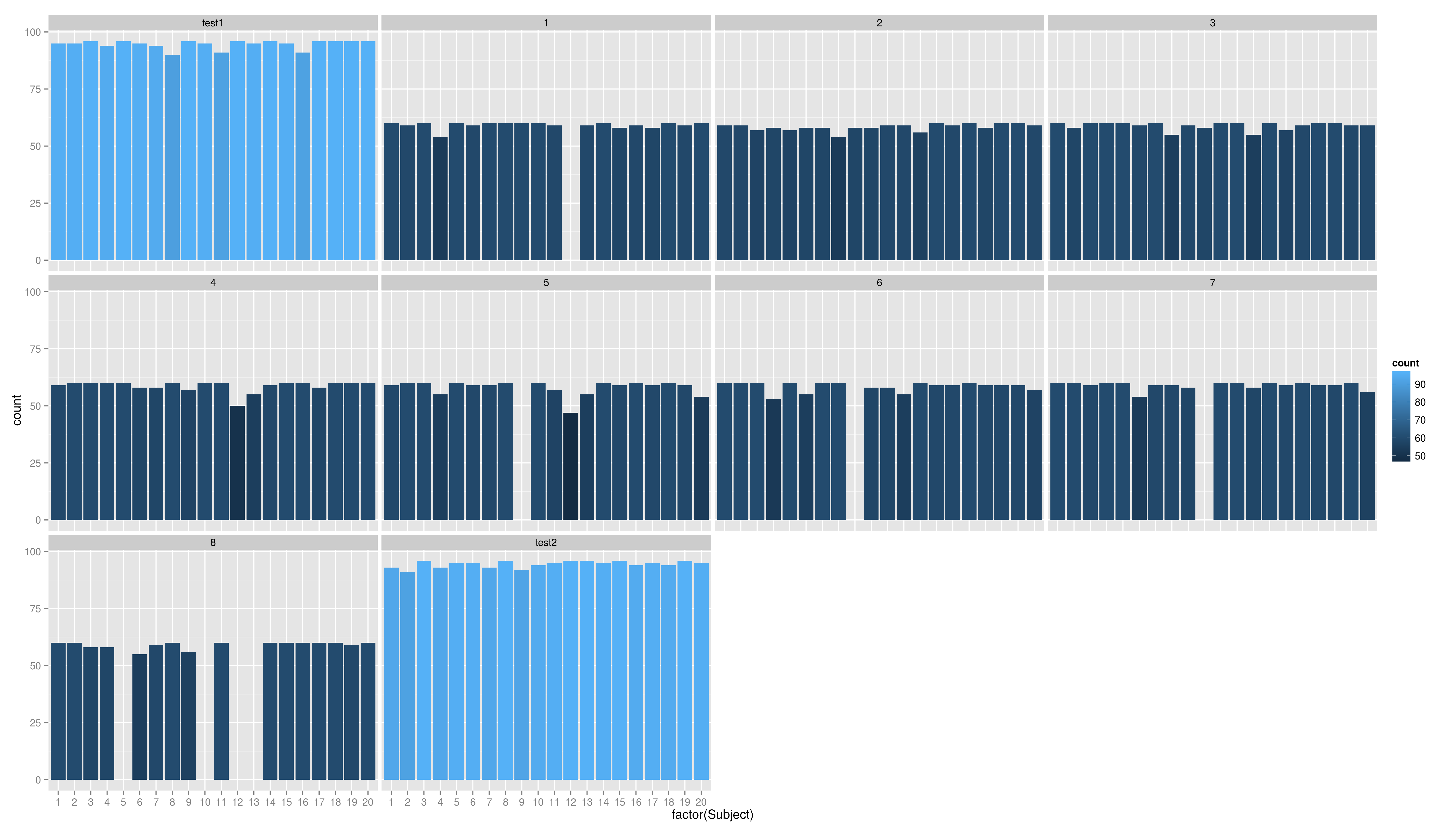
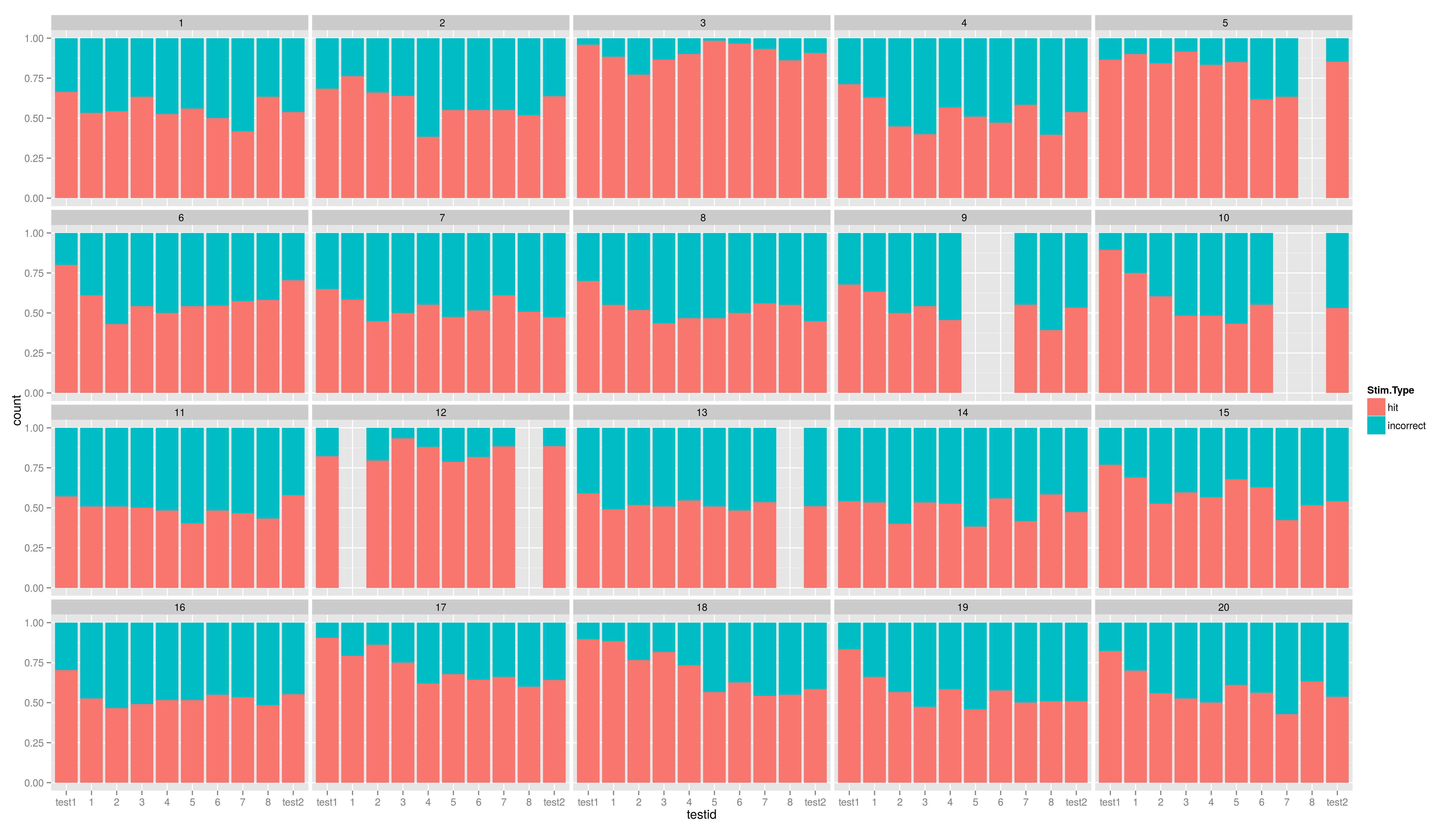
Just run the code and try to understand it. We will cover the ggplot graphics soon.
Summary Graphics
- so there are problems in coding of the test id
- we remove the letters at the end using str_replace()
1 > data$testid <- str_replace(data$testid,"[a-z]$","")
2 > data$testid <- factor(data$testid,
3 + levels=c("test1","1","2","3","4","5","6","7","8","test2"))
4 > table(data$Subject,data$testid)
5 test1 1 2 3 4 5 6 7 8 test2
6 1 95 60 59 60 59 59 60 60 60 93
7 2 95 59 59 58 60 60 60 60 60 91
8 3 96 60 57 60 60 60 60 59 58 96
9 4 94 54 58 60 60 55 53 60 58 93
10 5 96 60 57 60 60 60 60 60 0 95
11 6 95 59 58 59 58 59 55 54 55 95
12 7 94 60 58 60 58 59 60 59 59 93
13 8 90 60 54 55 60 60 60 59 60 96
14 9 96 60 58 59 57 0 0 58 56 92
Summary Graphics
Summary Graphics
And another one.